One of the most exciting uses of machine learning is scientific discovery. Sometimes long before a discovery is officially made people have written about similar ideas and one could speculate what might have happened if some dots had been connected sooner.
This is exactly what the mat2vec model (code, paper) seeks to do with the help of AI. It is a word embedding model trained on materials science literature that is able to capture latent knowledge. For example they show that you can predict material properties like thermoelectricity based on similarity of word embedding of “thermoelectric” to embeddings of different materials.
Another project HyperFoods uses graph neural networks with goal of identifying food-based cancer-beating molecules.
Inspired by these projects I hacked together veg2vec, a word embedding model training on scientific abstracts about health effects of different plant-based foods.
You can try out a demo of the model here.
Contents
- Veg2vec
- Food embeddings
- Food similarities
- Food molecule similarities
- Food health effect similarities
- Next steps
Veg2vec
Steps for developing the model
- I retrieved abstracts from the PubMed API by searching using as keywords various plant-based ingredients
- Then, with some help from a family member, I annotated about 1000 of the abstracts similar to the approach in the paper as relevant if about plants and human health or diet and not relevant otherwise (with a small number where it was doubftul)
- Neither of us are subject matter experts so although often it seemed quite obvious that an article, a scientist in this field might have labelled some of them differently.
- Then I trained a simple logistic regression model using TF-IDF features with 5-fold cross validation and a small test set on which it got an f1_score of 0.8
- I then used the model to filter the abstracts from over 520k to around 128k.
- Then I modified the
gensimword2vec model using in mat2vec and trained it -
I evaluated it on the grammar analogies they had provided and made some new ones, for instance one where common names of ingredients are paired with scientific names for example common name to species like
asparagus - asparagus_officinalis + brassica_oleracea = broccoli - The scale of the accuracies is lower to those presented in the mat2vec paper suggesting that the analogies are not as well aligned with the dataset and / or that the dataset could be improved
UPDATE
After some improvements including a more extensive list of n-grams, use of the preprocessing methods from the paper and more abstracts, results have improved as shown below. The improvements are quite signficant but the drop of about 0.25% for gram5-present-participle which accounts for over a third of the analogies which affects the total score. I have updated the tsne embedding plots although they don’t seem to look too different from before. Have yet to repeat the analysis for the other sections.
| relationship | accuracy-original | accuracy-improved | |
|---|---|---|---|
| 0 | common-family | 0.252381 | 0.412088 |
| 1 | common-genus | 0.019231 | 0.106061 |
| 2 | common-species | 0.318182 | 0.325758 |
| 3 | family-genus | 0.125000 | 0.178571 |
| 4 | gram2-opposite | 0.339827 | 0.372294 |
| 5 | gram3-comparative | 0.457895 | 0.536842 |
| 6 | gram4-superlative | 0.345455 | 0.454545 |
| 7 | gram5-present-participle | 0.220816 | 0.218306 |
| 8 | gram7-past-tense | 0.256354 | 0.296337 |
| 9 | gram9-plural-verbs | 0.478333 | 0.537931 |
| 10 | total | 0.274692 | 0.302738 |
Food embeddings
Ideally the word embeddings for foods should be close if they have similar nutritional properties and health effects but it is also expected that they will group in other ways
Before we look at results, please note:
- I am not an expert in health, nutrition, plant science and related so there are likely to be mistakes.
- Results based on co-occurrence of words in the dataset and must not be considered to have scientific validity - do not rely on them in any way!
Presence of cancer beating molecules
- I found a table which I understand is based on the HyperFoods project which contains the common and scientific names for several plant-based ingredients and for each one a list of the cancer-beating molecules found in them I augmented with food groups from FooDB
- To match the scientific names with the words in the vocabulary, I used
difflib - This method matches words using an algorithm that tries to ‘compute a “human-friendly diff between two
sequences’ according to the documentation. Sometimes the match is not right if for instance a similar enough result does not exist in the vocabulary. For example
actinidia chinensis(kiwi) was not present as a single ngram and this was matched toschizandra chinensis(five-flavour berry) - In the results below I filtered matches where the scientific term did not match correctly or where I was unsure if the match was correct
- In the plot below, the vectors for the scientific names are used and the size of the points is based on the number of CBMs present in an ingredients according to the dataset
- Evidently the ingredients cluster based on food group but there is also a suggestion that within a group those with similar numbers of CBMs are closer together
Alklaine diet foods
- Now we consider the foods show in this chart

- The TSNE plot shows ingredients which are present in the vocabulary, with the size of the points determined by the the alkalinity (largest for most alkaline and smallest for most acidic)
- For this plot, the common names are used after mapping to the vocabulary (as described earlier).
- The foods seem to cluster by category and there is some suggestion of subclusters of similar alkalising ability, which is more evident when coloured according to this feature.
Food similarities
Closest ingredient matches
Let us now look at which ingredients are most similar to a couple of different items using both the scientific and common names: celery and apium graveolens, cranberry and vaccinium macrocarpon
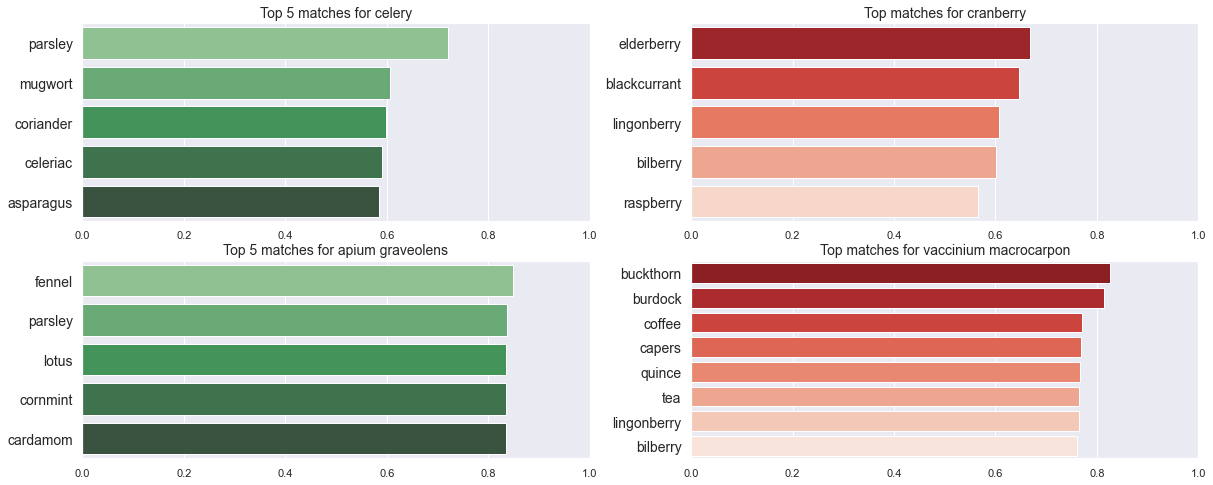
Observations
- Interestingly the similarities are much larger using the scientific name
- Moreover whilst all the common names co-occur with
celery, a simple co-occurrence test described below fails forlotus,cornmint,cardamom(nelumbo nucifera,mentha arvensis,elettaria cardamomum) andapium graveolens. Howevercelerydoes co-occur withcardamomandlotus. - For
cranberrythe most similar words to the common name are berries with which I am familiar but when using the scientific name such berries appear only below the top 5 - I was not familiar with buckthorn but turns out that the variety in the vocabulary (sea buckthorn or
hippophae rhamnoides) has edible berries
Finding similar local ingredients
Often data about ingredients is restricted to those available in some part of the world. Although the HyperFoods dataframe contains many fruits and vegetables from all over the world, what it does not include are what one might term “hyperlocal” :) ingredients, those which are not regularly eaten or readily available everywhere. I might know that kale has some beneficial properties and wonder which of several Indian green vegetables makes the best substitute - with regard to health and nutrition rather than taste. Here we rank these by cosine similarities of the scientific names of the greens and the brassica oleracea species which includes kale, broccoli, cabbage and other vegetables known for their health benefits.

Food molecule similarities
- I augmented the CBMs for each food in the Hyperfoods data using data from FooDB. In this preliminary analysis I only did this for a selection of CBMs (25 most frequently occurring and 25 with the highest similarity to the word
anticancer, then filtering to restrict to those present in the vocabulary). -
I was interested to know the following
- How many of the most similar ingredients to a given CBM have been identified in the dataframe as containing the CBM?
- How many of top matches co-occur with the CBM in an abstract?
- To approach the second question I used an estimate by looking at only those abstracts that had been retrieved from PubMed using the common name as a query keyword. For example to estimate if
quercetinco-occurs withvaccinium myrtillus(bilberry) in the text I looked through all the abstracts that had been retrieved by using the query keywordbilberry. This of course is only an approximation since the terms might co-occur in abstracts retrieved using a different keyword but the method was chosen in the interests of speed in this initial analysis.
Examples
- The top 10 matches for
quercetinamong ingredients in the CBM dataset areonion,grape,sorrel,guava,ginkgo biloba,bilberry,spinach,loquat,okraandblueberryall of which are known to containquercetinand this also co-occurs with all these in the abstracts dataset. The similarity scores are quite low however with the highest (onion) having a score of 0.21
- A less frequently occurring CBM is
betulinic acidwhich, according to the dataset, is found ingrape,rosemary,olive,pomegranate,basil,elderberry,chicory,walnut,persimmon,plum,abiyuch,jackfruit,nance,winged_bean. The most similar arepersimmon(I think since the matched word isdiospyroswhich is the genus),mangosteen,pepper,azuki bean,sapodilla,jackfruit,elderberry,mango,beetrootandlotus, which only contains 3 of the ones in the datasetjackfruit,elderberryandpersimmon
- The CBM
tricetinhas the highest similarity score toanticanceramong the CBMs in the table (0.6) and the top 10 matches arecherry,beetroot,gooseberry,rubus(which includesblackberryandraspberry),currant,winged_bean,sapodilla,capers,melon,star fruit. - The similarity scores for the ingredient matches are also quite high compared to those for other CBMs (the score for the highest,
cherryis 0.678 and that for the lowest among the top 10,star fruit, is 0.511). - None of the top 10 are in the CBM dataset. I had run queries only for
cherry,beetroot,gooseberry,currantandmelonas well asraspberryandblackberry(forrubus) and for all of these the co-occurrence test fails. In fact the word is entirely absent from all the abstracts queried with these keywords. The first one for which is succeeds isfenugreekwhose score is 0.505. - For the best match,
cherry, I ran queries across all abstracts for different scientific names forcherryandtricetinand there were no co-occurrences. - The two tricetin-containing foods in the dataframe are
gingko bilobaandtea. I did useteaas a search keyword butgingko bilobafails the co-occurrence test. The similarities for these are 0.314 fortea(camellia_sinensis) and 0.08 forgingko biloba- which might be because my retrieval was not comprehensive enough or because abstracts got filtered in the initial stage.
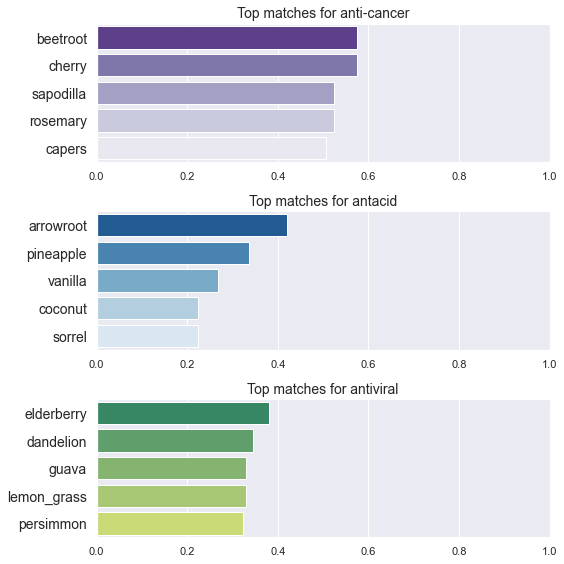
Food health effect similarities
- Let us now see what foods are most similar to terms related to health effects
- Since we have been looking at foods with regard to cancer and acidity, we will consider the terms
antacid,anticancerand for good measureantiviral - The vocabulary also contains
anti-cancerandanti_cancerso we find the similarity to each of these and use the max and forantiviralwe also useanti_viral
Next steps
- There are lots of improvements that can be made
- Better methods to evaluate whether latent knowledge has been captured
- A cleaner dataset addressing some of the issues discussed above
- Data cleaning and normalisation to get better embeddings for scientific names and names of chemicals. I provided a small list of scientific names of ingredients to use as n-grams but this could be improved.UPDATE: adding more n-grams and using the processing pipeline in Mat2vec (even though it was designed for papers about inorganic molecules) leads to significant improvements (see above).
- Extensions
- Training models for abstracts for published before the earliest co-occurrence of some words and evaluating the similarities to predict relationships
- Identifying other tasks for using the word vectors. I tried training models (logistic regression, SVM, small neural network) to predict number of CBMs and alkalinity but these did not yield good results.